Phytoplankton fluorescence and shape#
For a more detailed description see: https://www.phenopype.org/gallery/projects/phytoplankton-fluorescence/
import phenopype as pp
import os
## my root directory - modify as you see fit
os.chdir(r"D:\science\packages\phenopype\phenopype-gallery_exec")
## my laptop has a small screen, so I use a smaller phenopype window
pp._config.window_max_dim = 800
Make phenopype project#
Create a phenopype project (remember, Project is used to both create new projects, and load existing ones). Different phenopype projects can be useful, e.g., to process different batches of images from field seasons, perspectives, etc,. It makes sense to create them side by side in a subfolder, which I call “phenopype”. Thus, my research projects often have the following directory structure (just my way of working - this is really totally up to you):
my-project
data # processed data (e.g., tables)
data_raw # raw data (images, videos, etc.)
phenopype # phenopype projects
phenopype_templates # phenopype .yaml config templates
scripts # python, R, etc.
[...] # manuscripts, figures, literature, etc.
proj = pp.Project(r"phenopype\phytoplankton-fluorescence")
## add all phytoplankton images from the data folder, but exclude fluorescence channels
proj.add_files(image_dir = r"data_raw\phytoplankton-fluorescence", exclude="FL")
## add the config template; provide a tag
proj.add_config(template_path=r"phenopype_templates\phytoplankton-fluorescence_template1.yaml", tag="v1")
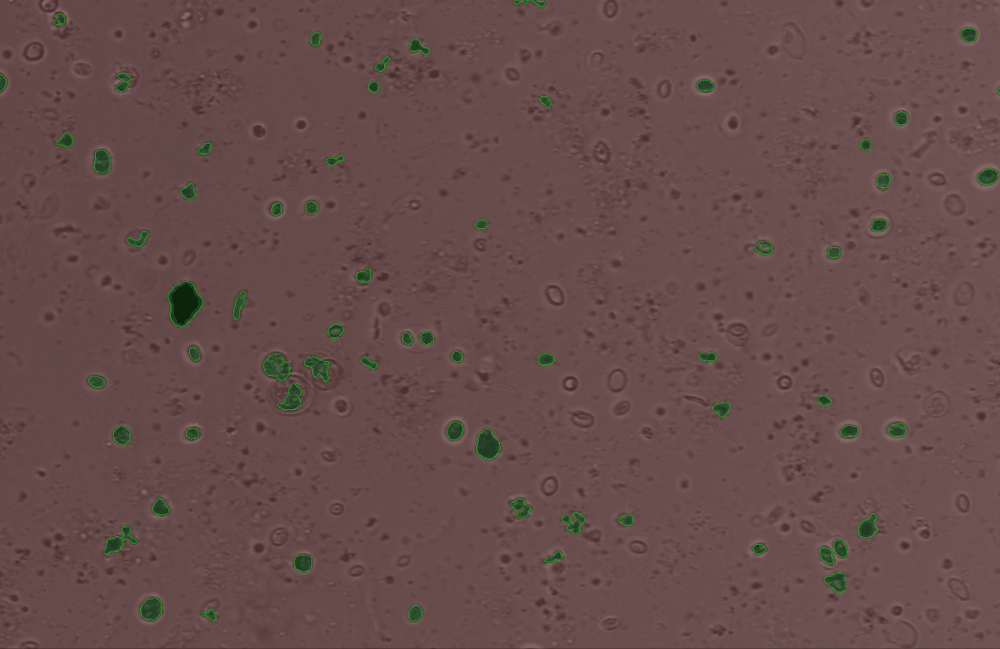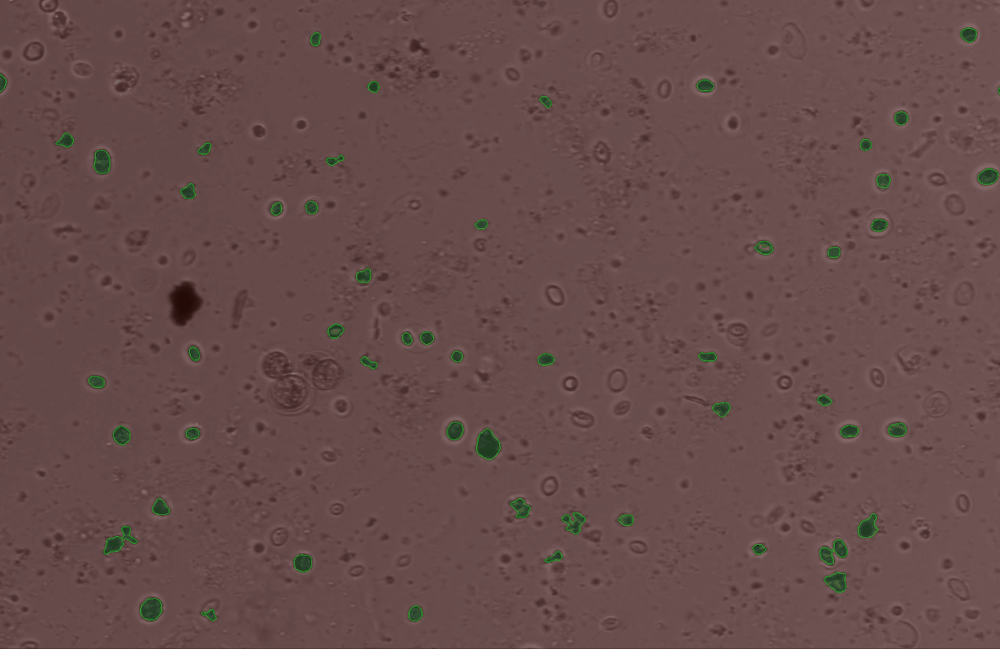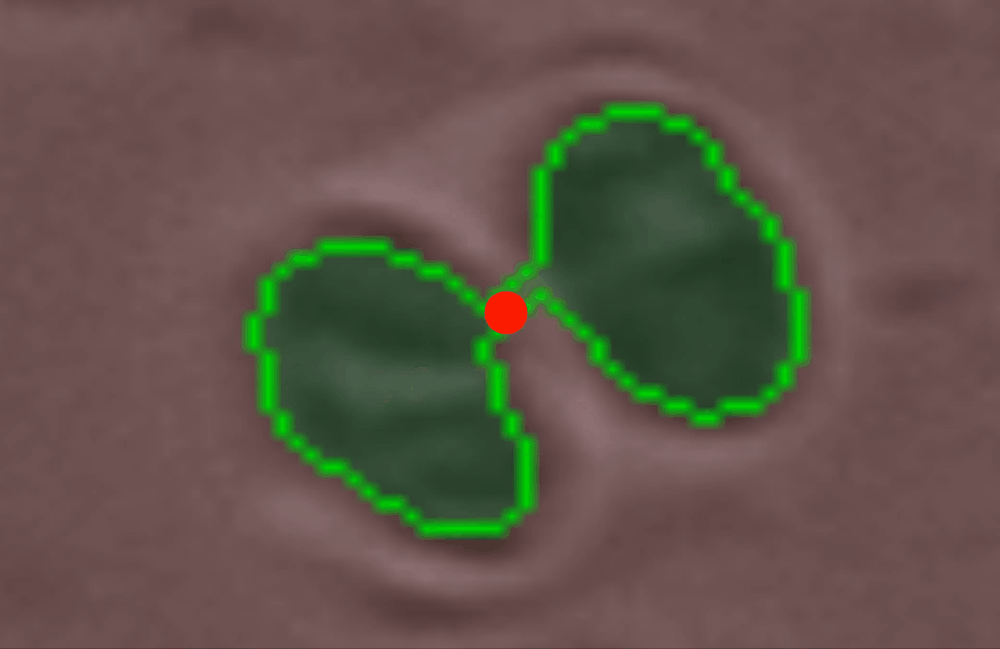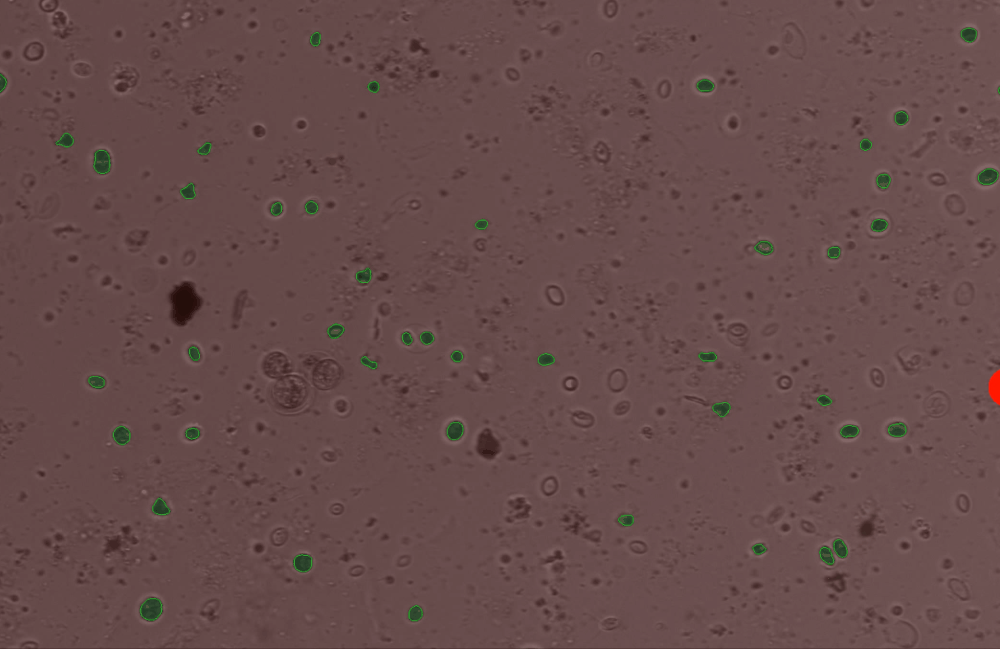
Use the edit_contour function to remove unwanted objects, e.g. cells that are broken, too small, or other unwanted junk and detritus.#
## run image processing (`window_max_dim` controls the window size of all GUI functions in the Pype config)
for path in proj.dir_paths:
pp.Pype(path, tag="v1", window_max_dim=1750)
Compute shape features of cells in brightfield images#
The shape features say something about cell morphology, which is why we only want intact cells. Since the fluorescence channels may not tell us whether a cell is intact or not, we only compute those features in the objects detected in the brightfield images.
## use `edit_config`´to inject `compute_shape_features` into the configuration files
## this makes the initial image processing faster, as this step is somehwat computationally intensive
target1 = """ - export:"""
replacement1 = """ - measurement:
- compute_shape_features:
features: ["basic","moments","hu_moments"]
- export:"""
proj.edit_config(tag="v1", target=target1, replacement=replacement1)
## run pype again, but without visual feedback to speed things up
## run image processing
for path in proj.dir_paths:
pp.Pype(path, tag="v1", feedback=False)
Compute texture featues of cells in fluorescence images#
This procedure uses the contour information we collected in the high-throughput workflow above. It provides all object coordinates to the compute_texture_features function, which, if also supplied with the fluorescence channel images, extrace texture featues from those coordinates. This code snippet shows that the low-throughput workflow, i.e., writing phenopype functions in pure Python code, can also have its use.
for path in proj.dir_paths:
## the _load_yaml function is part of the private API, and used here to load the attributes file to get the image name
attributes = pp.utils_lowlevel._load_yaml(os.path.join(path, "attributes.yaml"))
image_stem = attributes["image_original"]["filename"].partition('_')[0]
## we load the annotations collection in the high throughput workflow above - we need the contour coordinates of each object
annotations = pp.export.load_annotation(os.path.join(path, "annotations_v1.json"))
## we now loop through the files in the data folder, which are named like the brightfield image, and load those images
for channel in ["FL1","FL2","FL3"]:
image_fluorescence_path = os.path.join( r"../../gallery/data", image_stem + "_" + channel + ".tif")
image_fluorescence = pp.load_image(image_fluorescence_path)
## using the fluorescence image and the contours, we can compute texture features for each object. this is somewhat computationally intensive
annotations = pp.measurement.compute_texture_features(image_fluorescence, contour_id="b", annotations=annotations, annotation_id=channel)
## we store the textures back to the annotations file
pp.export.save_annotation(annotations, dir_path = path, file_name="annotations_v1.json")
proj.collect_results(files=["canvas", "shape", "texture"], tag="v1", aggregate_csv=True, overwrite=True)