Phytoplankton fluorescence and shape#
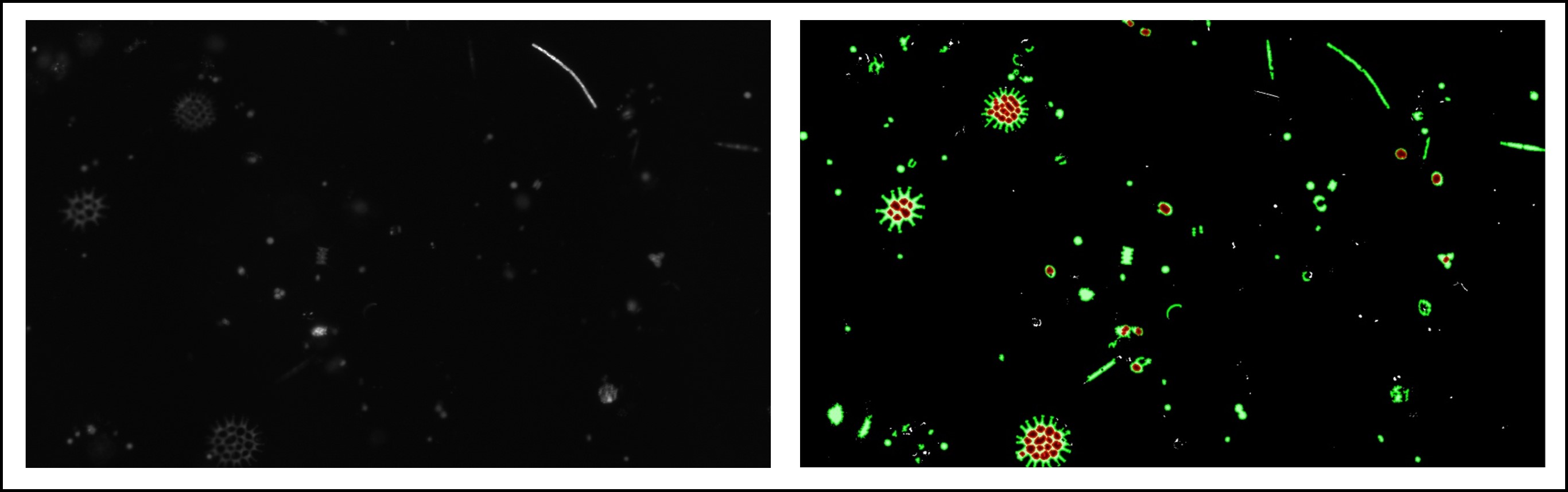
The goal here is to measure the density and distribution of photo-pigments inside individuals cells of phytoplankton, first performing thresholding to segment brigthfield images, and then use those masks and quantify pixel intensities in the fluorescence channels within.
Get started#
Read the jupyter notebooks for this project
Download the materials (see downloads section below)
Run the project yourself (see general instructions)
Background#
Competition for limited resources is a major force in structuring ecological communities. Cells belonging to different phytoplankton species within freshwater communities compete for light and nutrients in the water column. Quantifying pigment density through fluorescence intensities can help reveal patterns of competition and provide insight about how freshwater phytoplankton communities coexist and evolve. This dataset was collected under a nutrient limitation scenario (experimental manipulation of nutrients).
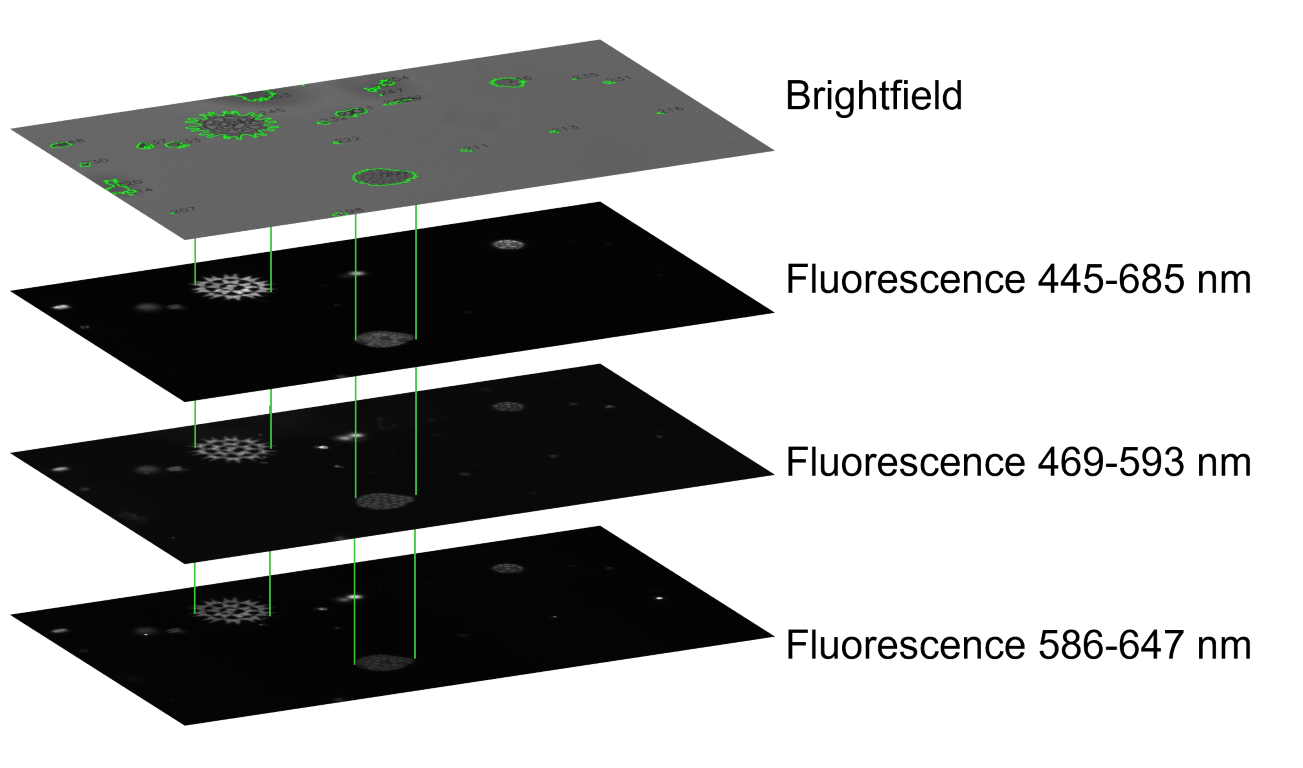
Each sample includes four images: 1 brightfield (top) and 3 fluorescence measurements (black, bottom).#
A plate reader creates four images of the same set of algae cells: bright field images to denote cell boundaries, and three different fluorescence channels to represent the distribution and density of different photo pigments inside each cell. All four images are single channel (gray scale) images. After segmenting the cells in brightfield, we can use the contour outlines as a stencil to extract texture information from the same coordinates in the fluorescence channels.
Jupyter notebooks#
Downloads#
Download data, scripts, and template