Cichlid teeth#
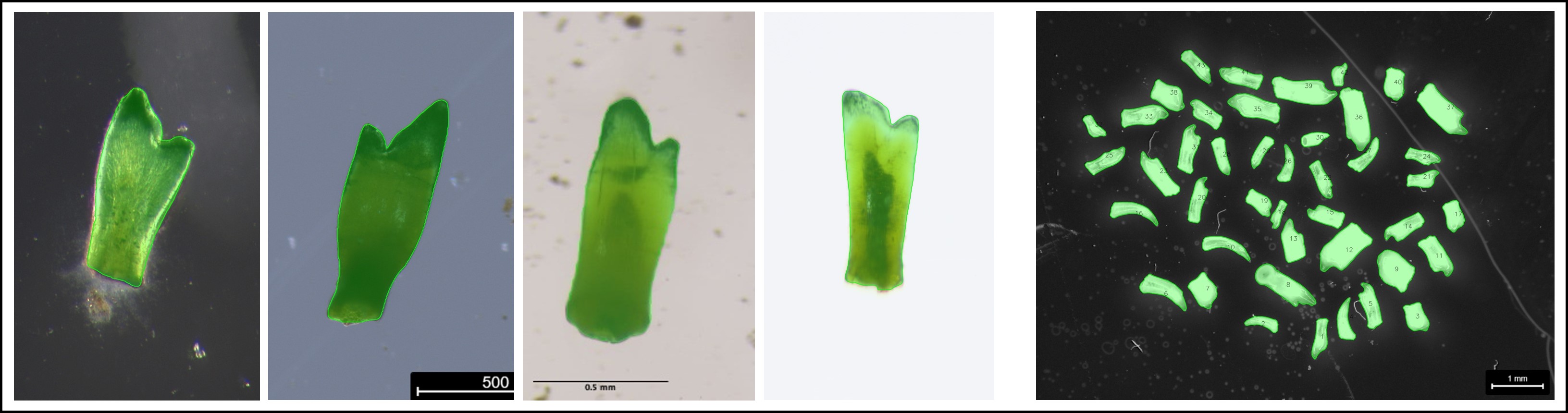
This example demonstrates how fish teeth are segmented from the background extracted from images containing single and multiple microscope images. The single tooth case is especially challenging because there are different backgrounds (light/dark) with various levels of noise.
Get started#
Read the jupyter notebooks for this project
Download the materials (see downloads section below)
Run the project yourself (see general instructions)
Install the fastSAM plugin for phenopype:
download the fastSAM repo:
git clone https://github.com/CASIA-IVA-Lab/FastSAMdownload a model checkpoint and place it in the fastSAM repo
pip install -r requirements.txt(not needed if torch is installed separately, which I strongly recommend - see below)install torch - go to the pytorch homepage and follow the instructions
install CLIP:
pip install git+https://github.com/openai/CLIP.gitinstall ultralytics:
pip install ultralytics==8.0.120reinstall phenopype (required after installing ultralytics):
pip install --force phenopype
Background#
These are single-tooth images from sediment cores that were photographed under the microscope. In these images, the background is sometimes brighter than the teeth, sometimes darker, and also somewhat noisy. This makes it hard for signal processing algorithms, such as the adaptive thresholding algorithm. This is an excellent opportunity to demonstrate the new fastSAM-plugin for phenopype, which uses mask as prompts for segmentation. The plugin is based on facebook’s Segment Anything Model, which is very good at detecting edges, so it should be able to deal with this variation. The fastSAM plugin is a much faster implementation of SAM that works very well on a local machine (detection speed with a GPU device is <1s).
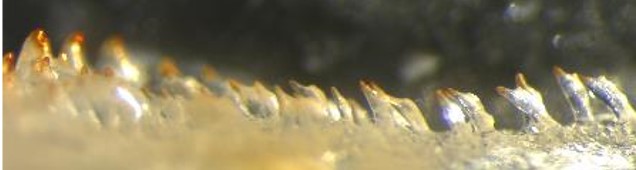
Multiple teeth are dislocated from a cichlid jaw to be imaged and used for a reference database of tooth traits.#
For the multi-tooth case, we can best use the adaptive thresholding algorithm, applied on the red channel where the teeth, which have a red tip, will occurr as all-white against a black background. This will lead to solid segmentation results.
Detecting the scale reference is a bit more challenging, for both the single- and multi-tooth case. In the single tooth case, I was successful with a configuration that picks up the completely black rectangle containing the scale:
## detect reference (single tooth case)
processing_steps:
- segmentation:
- threshold:
method: binary
value: 1
invert: true
- detect_contour:
min_diameter: 100
- export:
- export_csv
- visualization:
- draw_contour:
fill: 1
line_colour: red
Jupyter notebooks#
In notebook 1, we first need to load the fastSAM repo to the Python path before loading phenopype so it is recognized and the plugin properly loaded (see instructions above).
Downloads#
Download data, scripts, and template